by Ben Novak
The extinction of the passenger pigeon has challenged our conceptions of nature for a century. The bird that was “too numerous to go extinct” disappeared in a decade, leaving only skins and mounts in museum collections and a few in private hands. If the most abundant bird in the world could collapse at the hands of mankind, then perhaps any species was vulnerable to our onslaught. The loss of this astounding and albeit “larger than life” piece of American wilderness impressed upon citizens of the early 20th century to conserve our wild places and the creatures within them; saving the buffalo, the antelope, the egret, the turkey, and more species became a paramount endeavor for our nation. The conservation movement finally gained forceful momentum from the realization that even the most common species can vanish forever.

Male Passenger pigeon specimen from the collection of the Chicago Academy of Sciences and its Peggy Notebaert Nature Museum
To this day we still understand little about why the passenger pigeon became such a mega-species, flocking in the billions. And while we have learned, after the last century of research into the history of its extinction, that direct human harvest and disturbance are responsible for the extinction, we still do not understand just how these components caused the collapse of the species.
Biologists have known for a long time that studying how species have gone extinct will provide crucial insight into preventing species from going extinct in the future and saving species “on the brink” currently. But not until a few years ago did we have the tool to study extinct species the way we study living species: by deciphering the code of their DNA. For the first two decades of developing techniques to study what we call “ancient DNA” (the DNA of dead and fossil remains) we only obtained fragments of genetic material. But the diversity of those tiny bits of DNA was growing, including DNA from the Tasmanian tiger, dodo bird, the New Zealand Moa, the mammoth, woolly rhino, saber-toothed cats, Egyptian mummies, and even Neanderthals. In 2002, two genes of the passenger pigeon opened the door to the prospects of studying the most severe extinction event of human history.
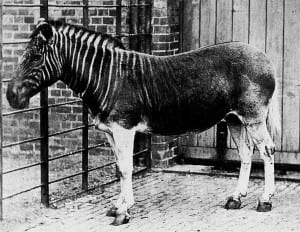
The quagga is a species of zebra that went extinct in the 1880’s. In 1984 it became the first extinct organism to have DNA sequenced, beginning the field of ancient DNA work and paleogenomics.
In the last five years technology and protocols have finally caught up with scientists’ ambitions. We have gone from sequencing small fragments of DNA to full mitochondrial genomes (17,000 base pairs of genetic code), and then in the last three years the field exploded into the realm of complete nuclear genomes. The Genome of the Neanderthal (3.3 billion base pairs) has been decoded from bones 30,000 years old. The “mammoth” genome of the woolly mammoth, at almost 5 billion base pairs of code, has now been fully sequenced and is soon to be published.
With the stage set for bigger research projects, many people began thinking about passenger pigeons. From autumn 2011 to spring 2013, independent research groups began studying the quality of DNA preserved in the tissue of taxidermy study skins of the passenger pigeon. From the first test sequences of 12 passenger pigeon specimens it was quickly realized that the full genome of the bird could be sequenced, and the exact way to do so became clear. With the small amount of test data alone the mitochondrial genomes of eight specimens have been fully assembled, and have directed insights into examining the past ecology of the species.
Simultaneously, advancements in other fields of DNA technology sparked the possibility of using the codes of genomes to engineer extinct organisms into existence again. Stewart Brand and Ryan Phelan founded the group Revive & Restore to use these technologies to bring extinct species back to life and to restore the genetics of endangered species. As they investigated what species could be studied for such a “de-extinction” project, the passenger pigeon came into the picture again and again from interviews with scientists. If the group wanted to bring the bird back to life, it would need to know the full genome sequence of the bird. It would also need to have the genomic code of a living pigeon in order to engineer the extinct code.
From the small fragments of DNA previously sequenced from the passenger pigeon, the closest living relatives of the passenger pigeon was identified as North American pigeons of the genus Patagioenas, including the west coast band-tailed pigeon. The new DNA from the test sequences have strengthened this conclusion, which came to a surprise to some ornithologists who had thought the mourning dove was the closest relative. As it turns out, the mourning doves and other American doves represent a unique group of columbiformes (pigeons and doves), which came from Asia separately from the ancestors of the passenger pigeon and band-tailed pigeon.
Early in 2012, I joined with Revive & Restore to pursue the goal of de-extinction via building a platform of genetic and ecological research. I had produced a portion of the test sequences mentioned earlier, with three specimens from a flock from New York in 1860, sampled from the collections of the Field Museum of Natural History, Chicago, Illinois. From the test sequence data, my colleagues and I knew that the specimens were “complex” enough to obtain the full genome, but the only way to put the code together would be by using the genome of the band-tailed pigeon as a reference map. The goal became obtaining band-tailed pigeon tissue for genome sequencing, which led to the partnership with Sal Alvarez, a private bird breeder with more than 30 years of experience working with North American pigeon species, including the band-tailed pigeon.
It was Dr. Beth Shapiro, an expert in ancient DNA and ancient population genetics at UC Santa Cruz, who first sequenced passenger pigeon DNA in 2002. Dr. Shapiro’s group produced the other portion of genome test sequence data, and from the variability of their results Dr. Shapiro knew that it would be possible that a new specimen might yield better data. The objective became finding that specimen. By 2012, Dr. Shapiro’s lab had obtained tissue specimens from 53 more passenger pigeons from the Royal Ontario Museum, Toronto, Ontario. With these additional specimens, we hoped to find the individual with the best quality DNA for the first full genome sequence. That first genome sequence would be used to assemble the genomes of multiple specimens, and the goal of studying the species’ evolution and ecology would commence – providing insight into the species’ life history and extinction.
It soon became clear that, although the goals of these research groups seem different, they are in fact convergent. Revive & Restore’s growing effort merged with the lab of Dr. Beth Shapiro, Associate Professor of Ecology & Evolutionary Biology at UC Santa Cruz in the spring of 2013. Work has expanded to include new tissue specimens from the Denver Museum of Nature in Science, Denver, Colorado, and to include bone samples from the archaeological collections of the University of Michigan Museum of Zoology, Ann Arbor, Michigan and the professional collection of Dr. Gregory Sohrweide of New York. To date DNA extractions have been performed on 56 tissue specimens and nine bones. The process for preparing these samples for genome sequencing has varied success with different DNA extractions. After library preparation of the samples, 55 of the total 65 had enough DNA to be indexed, or “bar-coded”, and amplified for sequencing. After the specimens were indexed, 34 of them have enough DNA to begin testing. These 34 samples represent 31 tissue specimens and three bones, dating from the 1600s to the 1890s from regions of Ontario, New York, Illinois, Michigan, to as far Northwest as the northern border of Minnesota and as far south as North Carolina. We will soon have our first look at the quality of DNA from these new specimens and the beginning foundations of our extensive work. We could have enough DNA sequence data to begin assembling the genomic code as early as December of 2013.
In order to assemble that code, we need the genomic code of the band-tailed pigeon. Blood tissue from “Sally”, a female bird of Sal’s charismatic flock has provided more than enough DNA to test new ways of preparing DNA for genome sequencing. Some DNA sequence data has already been produced, and the bulk is now in queue for sequencing at a larger scale. The band-tailed pigeon genome will be the second pigeon genome to be sequenced since the release of the rock pigeon genome earlier this year. Pigeon and bird genomes (including to date the chicken, turkey, and zebra finch) have averaged 1.2 billion base pairs. With the smaller size of these genomes, and the way they have evolved, the assembly of the band-tailed pigeon genome will not take long. Once the genome sequence is complete (late 2013) and finalized (sometime in 2014), major research into the evolution of the band-tailed pigeon and passenger pigeon will unfold, providing the platform for understanding how to recreate the passenger pigeon.
While this DNA work has been building, the ecology of the passenger pigeon has been going through a revolution of retro-analysis. The abundant historical accounts and records of the bird are being incorporated with newer data of the paleo-ecology of North America’s forests and the dietary ecology roles of pigeons around the world. Applying all of this to the passenger pigeon is challenging many widely accepted notions about the bird. As these new ideas about the passenger pigeon are discussed among conservation, management, and ornithology researchers there is growing agreement that the passenger pigeon is an excellent candidate for de-extinction. The next steps in this research are hoped to include experiments in recreating passenger pigeon ecology and thereby examining the actual effects the bird had on forest biodiversity and nutrient systems. Holland Shaw, a passenger pigeon enthusiast, is teaming up with Revive & Restore to assist in these experiments. Other projects are “in the works” in these discussions, and should have some cool insights in store.
In the next few years, conservation biologists and ornithologists will be able to utilize major knowledge gained from the study of the genome of the passenger pigeon for their own work with saving other species in danger of extinction, ranging from the knowledge gained to the biotechnology tested. And soon the field of conservation will expand beyond the re-introduction of locally-extinct species and the re-introduction of captive-bred species that became extinct in the wild to the actual re-introduction species that have gone completely extinct. In this way, the continuum of de-extinction efforts to conserve the value of ecosystems everywhere will unfold, illuminated on the blue wings of passenger pigeons.